
Leopold & Rudolf Blaschka, Doliolum Mulleri, National Museum and Gallery, Cardiff. Glass c1890, Of the genus of the Doliolidae, a free-swimming tunicate with a barrel-shaped transparent body. http:designmuseum.org/design/leopold-rudolf-blaschka
I began using the Department’s Drishti software in 2006. As I learnt to control a 3D object in a virtual space I experienced a similar feeling to blowing or turning hot glass on a punty; I could see the object but couldn’t touch it. Playing with my first dataset of a miniature plastic octopus, and exploring the variations of transparency and opacity, reminded me of the exquisite glass models of sea animals that were produced by Dresdon artisanal glass makers Leopold Blaschka (1822 – 1895) and his son Rudolf (1857-1929). In their lifetime the Blaschka’s supplied natural history museums across the world with thousands of realistic botanical and marine specimens. Their work was inspired by the rapidly expanding business of exploration and collection of rare and exotic species from both land and sea. The Blashka’s pieces are meticulously detailed and anatomically precise, and through the medium of glass they were also able to capture the translucence of creatures such as a live jelly fish. At the time natural history museums such as the Harvard Botanical Museum and the National Museum of Wales in Cardiff, commissioned the Blaschka’s to create models because they preferred these replica’s to the endless displays of grey lifeless specimens in jars.
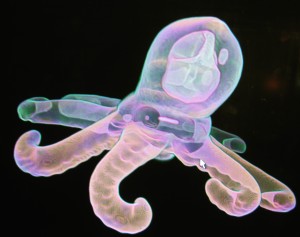
plastic octopus in Drishti, my first view 2006. Erica Seccombe, Vizlab, ANU Department of Applied Maths
It was not such a giant leap to make this connection as Drishti is very much in the same business as the Blaschka’s were. Drishti is a unique purpose designed volume exploration software so named by its creator Dr Ajay Lamaye, meaning ‘insight’ or ‘vision’ in Sanskrit. This open-source software is continually rewritten and upgraded with new features and was developed specifically for the end-use of visualising tomography or electron-microscopy data, for both exploration and presentation. Its premise in visualisation datasets acknowledges that the information is equally important as conveying understanding to a research community or a lay-person. Importing raw datasets into Drishti enables the user to identify the essential material by trimming a clean sub-volume for rendering. The program’s transfer functions distinguish between the material density of surface and sub-layer data through the process of elimination or enhancement, for example; separating nerves, veins, tissue and bone. There is a wide range of choice through colour, rotation, cropping, clipping, positioning and fly-throughs, with the additional function of a key frame editor to capture still images and create animations of any length.
Since its inception in 2004, Drishti has improved considerably in terms of its user interface, but as a user I have discovered that visualising true volumetric data is an acquired skill. The ability to image a dataset is not a straightforward procedure if you are unfamiliar interpreting volumes though histograms or navigating around a virtual 3D environment. But the point of end use is that you don’t have to be specialised in visualisation or skill, in the way that Leopold and Rudolf were dedicated to creating and constructing intricate models out of glass. But as a non-scientist and non-computer programmer I have been on a steep learning curve ever since I started my project. As I have mentioned before in previous postings, there is a great advantage of working in Vizlab and having Ajay just down the hallway. Many a time I have knocked on his door and he always has time for me, no matter how trivial the problem. If I didn’t have Ajay there to demonstrate functions or sort out glitches I would be lost. I do pride myself in being an independent user of software programs so when I’m using Drishti I always refer to the user’s manual on line. But I know when to call on Ajay.

Ancient Gogonasus advances evolution, Museum Victoria. The assembled complete skull of a gogonasus, whitened with ammonium chloride. Image: John Long, Museum Victoria
In 2006 there weren’t other people around actively using the program but as the 3D technology is now in demand, I am beginning to see how researchers are utilising visualisation tools for specific projects. I feel it is important for me to look at how data is treated through research as it take me out of the artist vacuum and in turn informs my work. For example here is a big difference between my datasets and 3D scans of fossils which are a billion times more detailed than a plastic miniature object or a mung bean. It is the scanning of fossils at the XCT facility and their visualisation in Drishti which has been one of the most interesting events. My basic undertanding of a fossil was that it was an imprint of the former being preserved for all time, and that the most solid examples of fossils are bones. What I didn’t realise was that fossils are often complete preservations, inside and out. Now that 3D microcomputed tomography is available these fossil specimens are now challenging the established understanding of evolution.
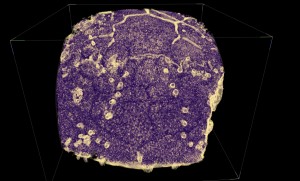
Gogonasus imaged in Drishti. Associate Professor Tim Senden and Dr John Long (Museum Victoria) By the time fish crawled up on land most of the big evolutionary steps towards humans had been made - some 350 million years ago and a good 100 million years before dinosaurs. ‘Gogonasus’ is one of the pivotal fish in this evolutionary picture. Discovered in the Kimberleys in June 2005 the complete, articulated and three dimensionally preserved skeleton gives us a remarkably detailed insight into how forelimbs and air breathing developed. It challenges the view that a single species made the transition to land and shifts the focus to Gondwanaland as another source of the necessary biodiversity to support tetrapod (all animals with four limbs) development. Three dimensional X-ray microscopy (X-ray CT) is also helping to probe the way the sensory system was arranged and the complete set of facio-cranal nerves have been mapped with this technique, and completely mirrors that in human skull. Indeed the pectoral fin is comprised of the same set of bones we have in our arms. The discovery of this specimen is significant in that builds a greater understanding of why some fish have remained fish and others started to sniff air.
Fossil hunting is a big business and the monumental finds have always rocked the establishment. I’ve always like the story of Mary Anning (1799-1847), her famous find as a child in Lyme Regis is now fictionalised in Curiosity by the novelist Joan Thomas. Still the most exciting event for a paleontologist is to discover a fossil in 3D entirety. Fossil preparation is a whole industry of its own and there are many slow and painstaking processes one might go through to extract a fossil from rock; such as acetic acid baths, penetrative plastic or resin coatings. Traditionally, to see the inside of a fossil it would have to be physically sliced up. If the fossil is destroyed in the process of examination there is no going back, particularly if it is a rare find. Prior to X-ray CT there was no easy way that such detailed structural information could have been extracted from a fossil. Through X-ray, the fossil can be scanned inside a lump of rock, the rock dissolved virtually though transfer functions in Drishti. Even if the original fossil is destroyed or lost the full detailed dataset remains as an electronic resource that can be turned, cropped and examined repeatedly.
Through the XCT facility there is a very specialised group of Paleontologists lead by Professor Ken Campbell (ANU), Dr Gavin Young (ANU), DR Kate Trinajstic (CUSTIN UNI), Dr John Long (LA County Museum) and Dr Richard Barwick (ANU) working with Tim SENDEN on scanning fossils of the Devonian lung fish. The term Devonian describes a geological period that existed over 400 million years ago and examples of life systems from this time show creatures adapted to various environments and climate, for example, fish evolving legs to crawl on land. These paleontologist are interested in these early vertebrates as our earliest known ancestors, and they have been mapping and comparing physical features to establish evolutionary theories. Micro-CT is providing an even greater level of clarity by revealing finer details of sensory systems inside the fine cranial and spine cavities, throut, snout, ears and jaws, soft tissue and even neurons, informing new research on the evolution of vertebrates. It is amazing to behold these scans in 3D. To the untrained eye they could be any bit of bone but Tim has pointed out to me the sections of fossil that are important.

The Queensland Lung Fish is one of three species of lungfish in the world and has a single lung-sac and breaths using a buccal force-pump similar to that of amphibians. It can grow up to 1.5 metres in length, weigh up to 40kg.
Dr Gavin Young warned me that if I am to mention placoderms I must not confuse the terminology with Pachyderm, a common trap for the unversed. Placoderms are considered the most diverse groups of the early jawed fishes. There are a recorded number of around two-hundred varieties of placoderms which evolved mainly during the Devonian period which diversify considerably but many becaming extinct at the end of that period. The ones that survived went on to develop into the first vertebrate giants. But I have learnt that the Queensland lungfish, Neoceratodus forsteri, is one of only THREE existing lungfish species left in the world and belongs to an ancient family of fleshy-finned fishes and is one of the oldest living vertebrates on the planet. Fossil records date this species back around 410 million years at the time other vertebrates were beginning to evolve and this species is often referred to as a ‘living fossil’ as contemporary specimens have remained unchanged over the last 100 million years.

John Long (left) and Tim Senden who found the specimen at Gogo 11 July 2005. Image Michael Nossal, Source Museum of Victoria
During the Devonian period the Australian landscape was vastly different to how we find it today. Apparently Canberra region was tropical covered in a shallow sea filled with coral reefs populated by lungfish. Wee Japser just west of Canberra is now a palaeontologists heaven for this period. Other sites range around the Kimberlies in WA in particular the Gogo formation inland off the coast, also a former reef, which is the only Devonian site in the world where you can find whole perfect, uncrushed fossils of fish. But don’t think you can just trip out there and find a few, its requires a lot of hard work and expertise; you would need to know what you are looking for. In 2005 Tim famously discovered the first complete fossil of well preserved Gogonasus. On his first field trip in the Gogo region with Dr John Long and his colleagues from the Melbourne Museum, he found one half of this fossil. When they realised how important this fossil actually was they returned to the exact spot according the GPS reading and Tim found the other half. This collaboration between Senden, Long and Trinajstic lead to another remarkable find of the oldest living evidence of live birth in a placoderm. By highlighting the umbilical cord and the baby fish in Drishti can the untrained eye understand what they are talking about. See the movie here. Naming this species Materpiscis attenboroughi after Sir David Attenborough, Senden, Long and Trinajstic launched their find at the Royal Institute Australia with a satellite hook up with Queen Elizabeth II !!
Quick links to these events, subjects, people & references can be found at:
RAPID ROUND UP: Worlds oldest Mother – live birth in the Devonian – experts respond
Ancient Gogonasus advances evolution, Museum Victoria.


2 Responses to inside out…